This page contains our current work and previous work. A list of our group’s scientific publications can be found here.
Current work


We are now recruiting all cause sepsis patients admitted to a general ward or to a high dependency/intensive care unit in Oxford, to this integrated, multi-omic study of the host response to sepsis. Samples are also being obtained from convalescent survivors.
Researchers: Andrew Kwok, Patrick MacLean, Hanyu Qin, Eddie Cano-Gamez, Justin Whalley, Yuxin Mi, Alice Trickett, Chloe Wainwright, Maddy Smee
Sepsis endotypes from transcriptomics
We generated genome wide gene expression data for peripheral blood leukocytes from 552 sepsis patients. Using this data, we identified and validated two distinct sepsis response signatures (SRS1 and SRS2) with pronounced differences in gene expression patterns and mortality rates.
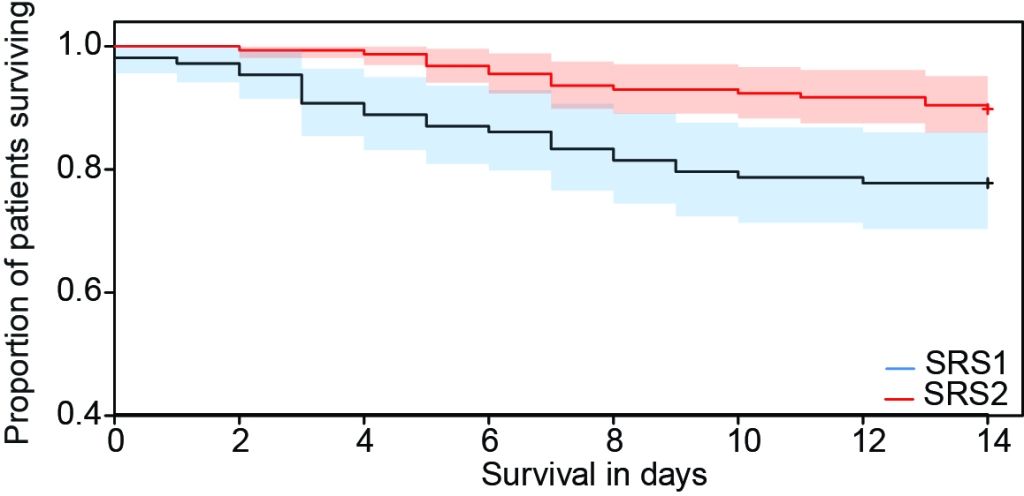
We also demonstrated that these disease endotypes may be resolved using a small gene set. These findings, which are supported by comparable results in other studies (e.g. MARS), highlight an opportunity for patient stratification and a precision medicine approach to the management of sepsis.
We have recently designed an R package containing a method for stratification of patients with acute infections using transcriptomics and applied it to sepsis, influenza and Covid-19. Researcher: Eddie Cano-Gamez

Since the start of the pandemic GAinS2 has been repurposed to recruit patients with COVID of varying severity. We have adopted an integrated, multi-omic approach to identify features of the immune response associated with COVID severity. We also compare the response to COVID with that seen in sepsis or influenza.
We have published a Multi-omic Blood Atlas for COVID (COMBAT) that defines hallmarks of disease severity and specificity. Researchers: COMBAT consortium

We are currently generating mass spectrometry proteomic data for a subset of patients enrolled to GAinS in order to further investigate heterogeneity in the sepsis response. Early results indicate that the SRS endotypes we describe in transcriptomic data are also reflected in the plasma proteome.
Researcher: Yuxin Mi

For more than half of patients recruited to GAinS with pneumonia, the causative pathogen was not identified using standard clinical tests. We are using next generation sequencing methods to detect and classify all genetic material present in the blood of sepsis patients, and thus improve knowledge of infectious aetiology in the GAinS cohort.
We have found evidence that Epstein-Barr virus reactivation was associated with the relatively immunosuppressed SRS1 endotype and differential expression of a small number of biologically relevant genes.
Researcher: Cyndi Goh

Patients and members of the public are integral to our research efforts. We have started a patient and public involvement project with 14 participants to improve the Sepsis Immunomics study. They are from all parts of the United Kingdom and they have been great help in planning and designing this website and the study.
Researcher: Angie Lee
Previous work
Expression quantitative trait loci
By analysing gene expression as a quantitative trait, it is possible to identify putative regulatory variants and their target genes. As the regulatory landscape is highly context specific, and known to be dynamic in the immune response, we mapped expression quantitative trait loci (eQTL) in sepsis patients and identified associations for a number of disease-relevant genes.
GAinS 1 study (complete)
Patients admitted to an intensive care unit (ICU) with sepsis due to community acquired pneumonia or faecal peritonitis were eligible for enrolment in the original GAinS study. For each patient, we obtained a blood sample for DNA extraction upon recruitment. On the first, third, and fifth day in ICU, we also took plasma and urine samples, and stored peripheral blood leukocytes for RNA extraction.
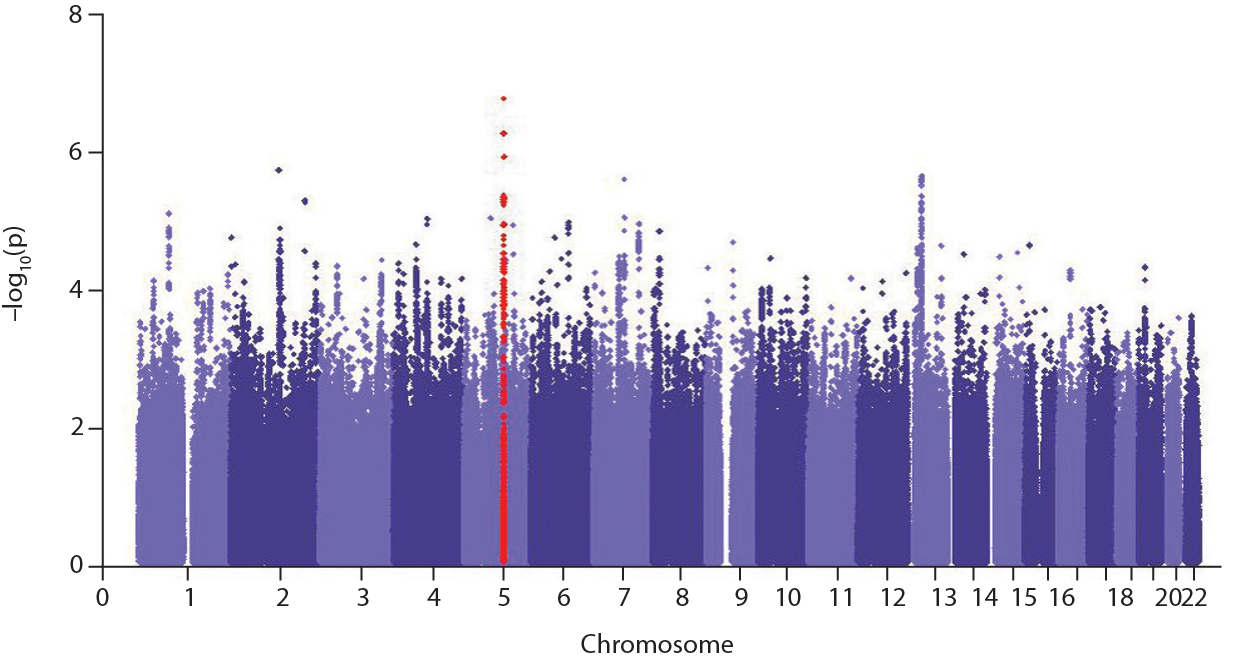
Genome wide association study for sepsis survival
Our genome wide association study for survival from sepsis was published in the Lancet Respiratory Medicine. This was the first GWAS for outcome performed in sepsis patients, carried out as part of the European GenOSept (Genetics Of sepsis and Septic shock) study. In a meta-analysis of 4 cohorts of pneumonia patients, we identified common variants in the FER gene associated with a reduced risk of death from sepsis.
Differential treatment responses in sepsis
In the context of the VANISH clinical trial, we found that the SRS endotypes showed different responses to hydrocortisone.

The 100,000 Genomes Project
The 100,000 Genomes Project is a flagship study that aims to sequence the genomes (the entire set of DNA in an individual) of 100,000 individuals in the UK. Only those with a rare disease or cancer are eligible for participation, since it is these conditions which have the most potential to benefit from this type of genomic study.
Although sepsis is a common condition, it can be considered a rare disease in some individuals who develop particularly severe sepsis despite having no risk factors (i.e. are young and previously fit). We have recruited a small number of such patients, under the age of 50, who had been admitted to an intensive care unit with severe sepsis caused by community acquired pneumonia. In such individuals, it is possible that a rare genetic abnormality contributed to the development of sepsis.
Recruitment has now ended and we are now in the sequencing and data analysis stages for this project.